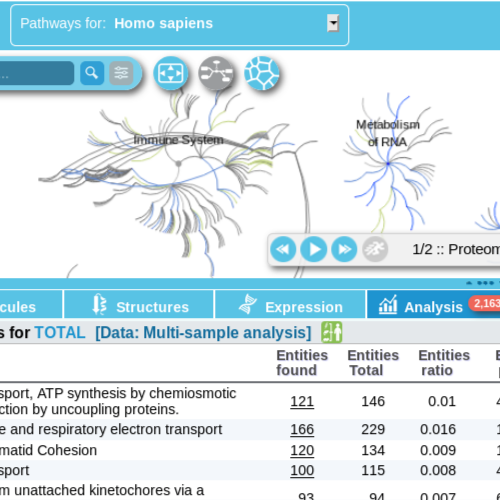
Reactome Pathway Browser
Web-based multi-omics cross-species pathway analysis
The Reactome pathway browser is the simplest approach to use the new ReactomeGSA analyis. Simply open the Reactome Pathway Browser and select Gene Set Analysis on the left. This will allow you to launch a new ReactomeGSA analysis.

ReactomeGSA
A Bioconductor R package
The ReactomeGSA R package is available on Bioconductor. It directly supports data structures from, for example, limma and edgeR which makes it straight-forward to perform multi-omics, multi-species comparative pathway analyses. Additionally, it has dedicated functions to perform pathway analyses of single-cell RNA-sequencing data.

Tutorials
Learn the ReactomeGSA R package
The quickest way to see the full potential of the ReactomeGSA R package is through our collection of tutorials. There we show how to use the ReactomeGSA R package to perform large-scale comparative pathway analyses of TCGA and CPTAC cancer studies and characterise single-cell RNA-sequencing data (go to tutorials site).

ReactomeGSA Backend
A kubernetes application
The complete backend of the Reactome Gene Set Analysis service is developed as one kubernetes application. It is designed so that it can easily be deployed on any kubernetes cluster. Therefore, for cases where the public analysis service cannot be used, it is straight forward to deploy one’s own analysis service. All details, as well as the source code are available at the project site.

Funding
This project has received funding under the European Union’s Horizon 2020 research and innovation programme under grant agreement No 788042.